

About lncRNA subtypes and ID assignment
|
|---|
1. lncRNA subtypes
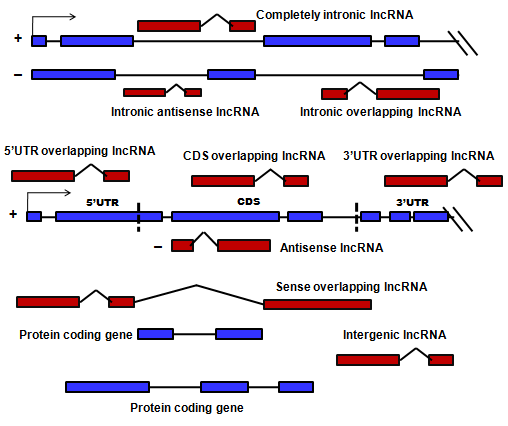
2. Assignment of lncRNA IDs
Based on the chromosomal location, we assigned unique identifiers to each transcript. Human lncRNA transcript identifiers (ID) begin with hsaLB. Human lncRNA transcripts have been designated a unique identifier as hsaLB_[type]_[numeral]. 'type' is the distinct subtype of the lncRNA transcript IDs. The numeral ({primary}.{secondary}) is assigned as per the chromosomal location of the transcript. Multiple transcripts from the same genomic locus have same {primary} number with increasing {secondary} number which represents different cDNA variants from the locus. Similar naming convention has been followed for mouse, drosophila, zebrafish, cow, chicken, rat, c.elegans lncRNA transcripts, preceded by 'mmu', ‘dre’, ‘dme’, ‘bta’, ‘gga’, ‘rno’, and ‘cel’ prefix respectively.
Search LncRBase v.2
|
|---|
1. LncRNA transcripts
5 search options are available in LncRBase v2 under Section Transcripts. 1. Search by Accession ID. First select the Organism name, then type a. LncRBase v2 ID to view detailed information on that transcript. or b. Original ID (Ensembl/Noncode v5.0/UCSC Gene ID/NCBI Refseq /H-InvDB 9.0). Clicking Submit displays detailed information about that lncRNA. 2. Search by Gene Symbol. To search for a specific lncRNA, first, select the Organism name, then type its gene name or gene symbol (HGNC/MGI/ZFIN/RGD/FlyBase/WormBase). You will be shown the number of entries for that gene in LncRBase and associated literature. Clicking on the particlar gene loci will show you the details of the gene and correponding transcript variants that are hosted in LncRBase. 3. Browse by lncRNA subtype, coding potential and sORF overlap. In the first segment, select organism first. Then select any specific lncRNA biotype, chromosomal position, coding potential tool name and coding potential type to view the corresponding information. For the second segment, organism and chromosome input would retrieve the sORF count of overlapped lncRNAs. 4. Search for associated genomic elements. First, select the organism. Then select either th Repeat/ CGI/ Microarray probes option and chromosome no. to view Repeat overlap, CGI in lncRNA promoters and remapped Affymetrix Microarray Probes respectively. 5. Browse small ncRNA associated lncRNAs. Firstly, select the organism. Then select either the miRNA or piRNA option and chromosome no. to view lncRNA-associated miRNA or piRNAs.
2. LncRNA expression
Click any of the eight (8) organism name under section “Expression” to open the organism-specific expression query page. Select a specific “tissue” to show all the corresponding data “accession IDs”. You can further filter your query by selecting type of lncRNA you want to search. In the output page you will be shown the total number of transcripts in that biotype and their average FPKM value in the selected tissue.
View results of your search
|
|---|
1. Detail Output of LncRBase ID Search
Clicking on an individual transcript ID in the general output page will direct you to a detailed output page. You can view LncRBase ID, Original ID, Alias ID , Gene name, genomic location, transcript length, %GC, Coding Potential score and predicted localization. To view detailed information on sORF count , associated miRNAs, associated piRNAs, number of microarray probes mapped to that lncRNA, predicted localization, associated Repeat Element, CGI, target genes, disease association and TFBS in lncRNA promoter well as lncRNA expression of various tissues, click on the view button.
2. ID Conversion
Select the organism( only for human and mouse) and input v2 ID to retrieve v1 ID if present. Click on the link to go to LncRBase v1 webpage of transcription ID details.