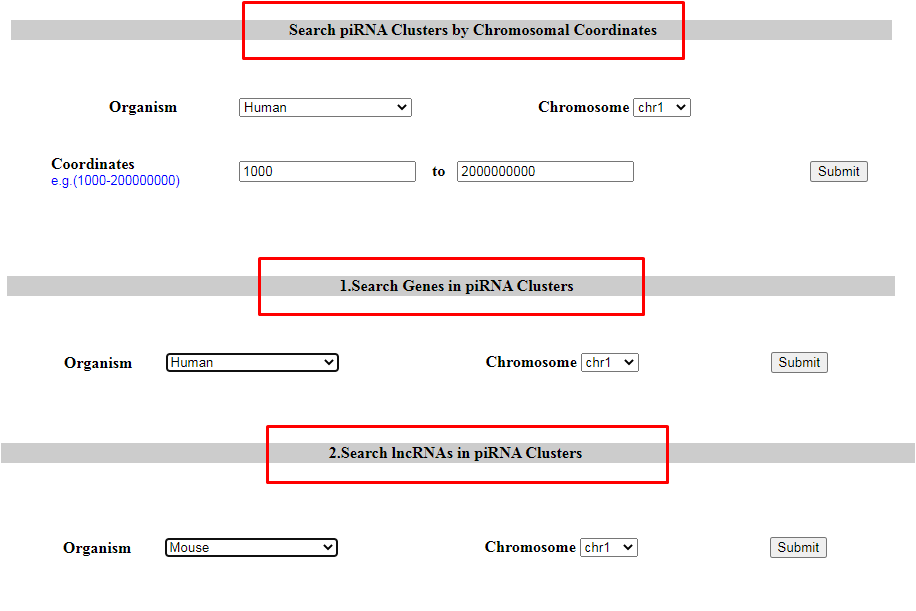
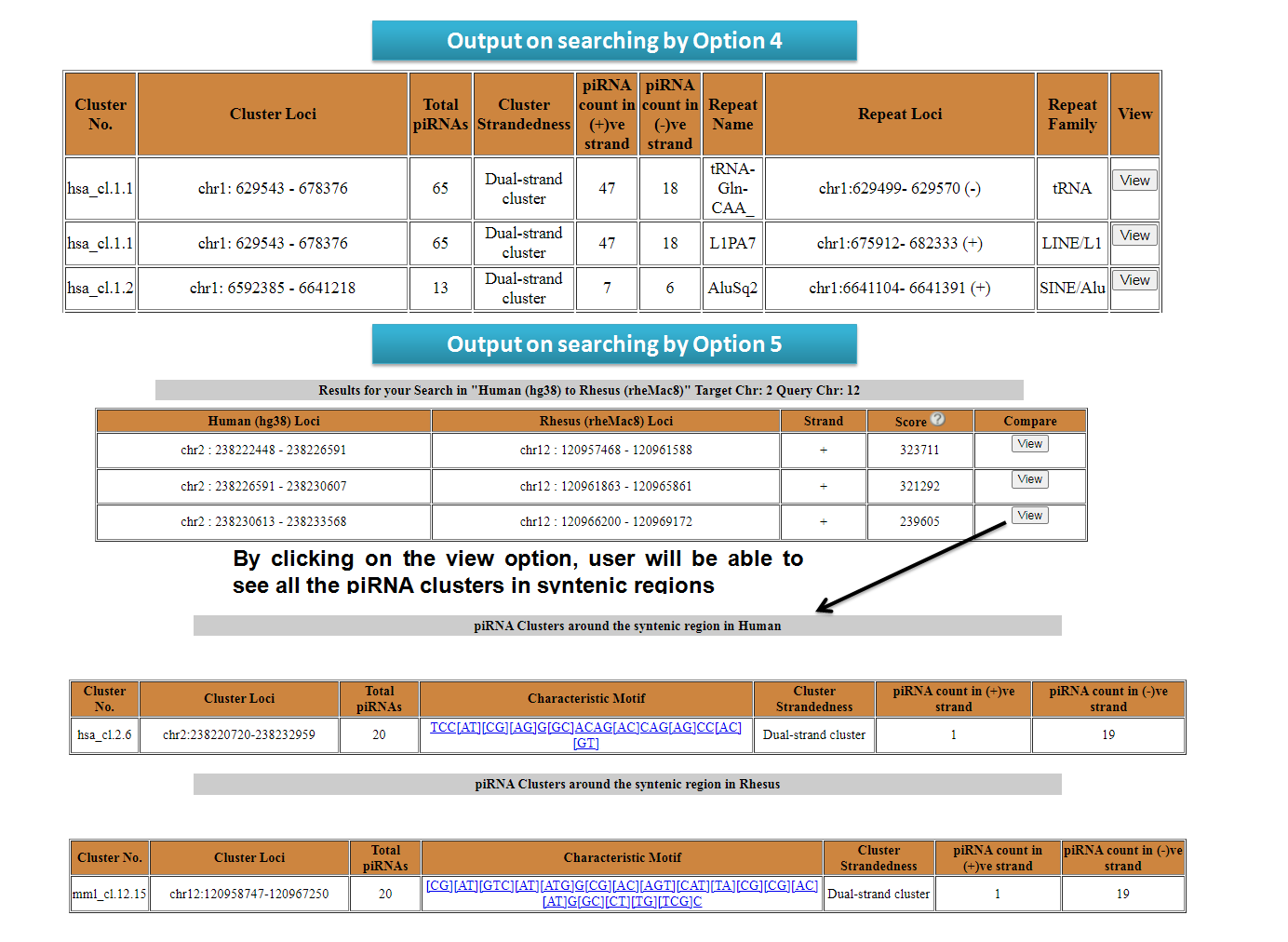
2. Search piRNA cluster
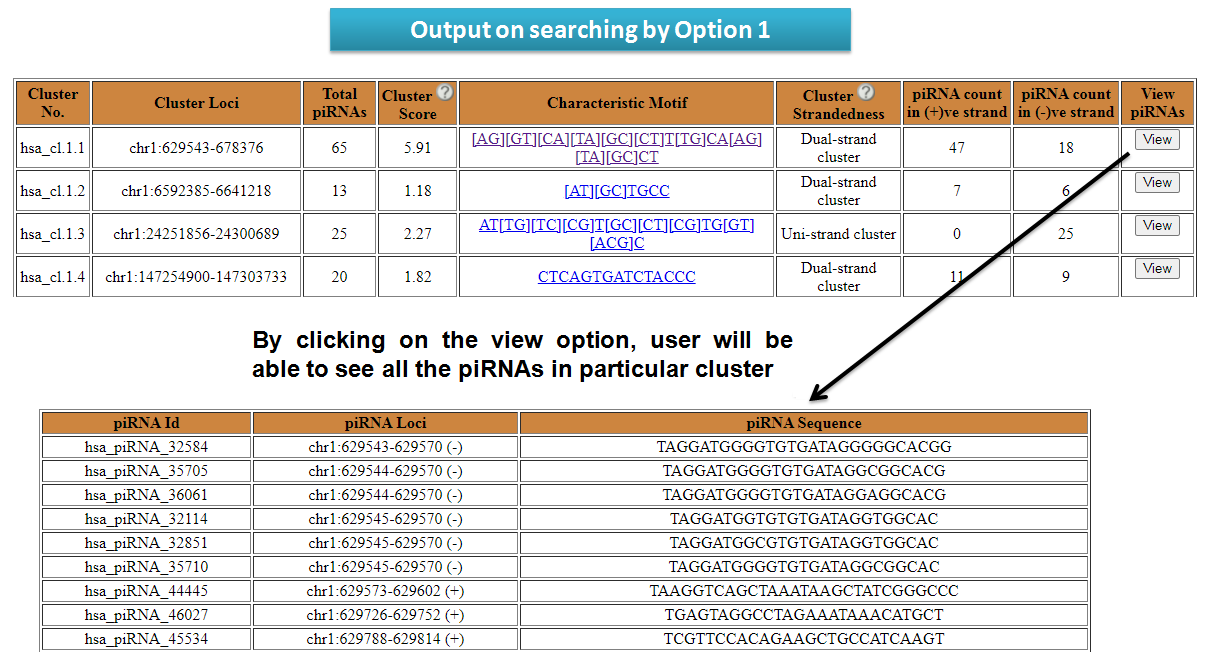
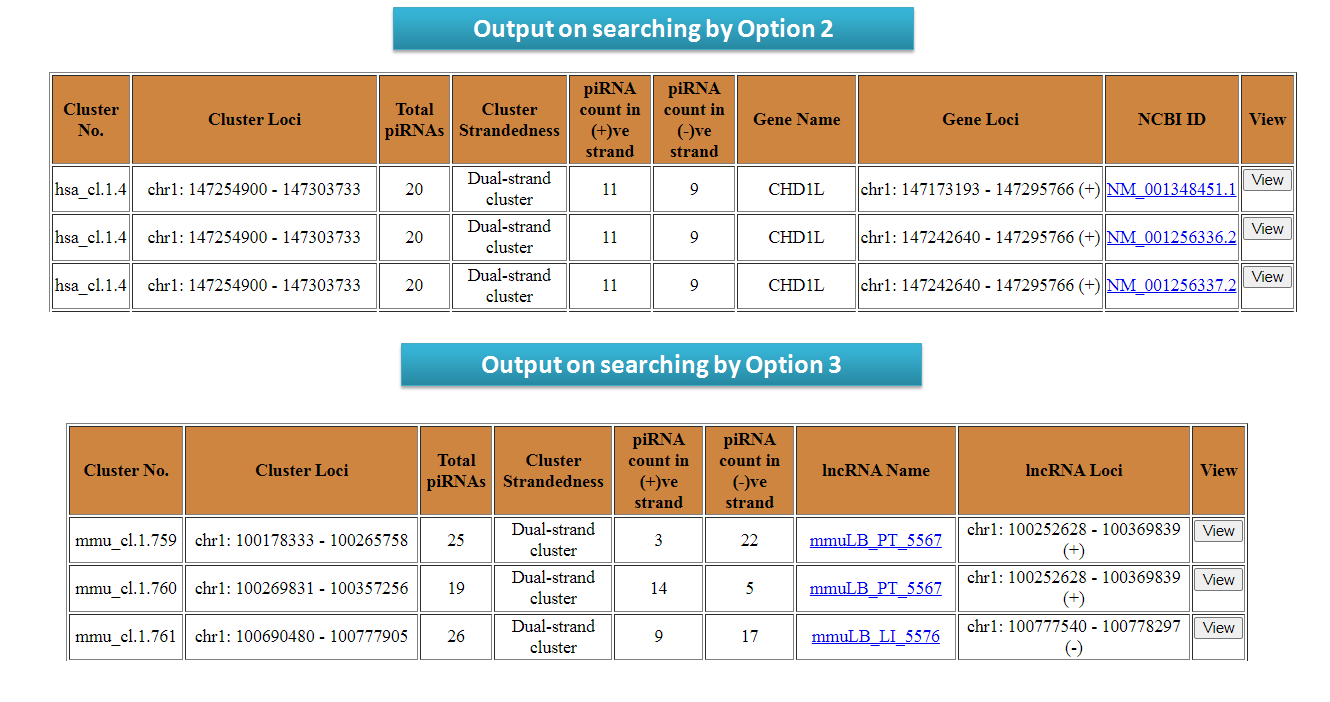
piRNA clusters can be searched by users by entering chromosomal coordinate (Option 1). One can also search for piRNA clusters present in genes (Option 2), lncRNAs (Option 3) and repeats (Option 4) as well. The output table will fetch detailed results of the piRNAs in repeat elements. Users can search for syntenic piRNA clusters by chromosomal coordinates. The output table will fetch the piRNA clusters corresponding to syntenic regions (Option 5).

View results for search options in piRNA Cluster
|
|---|
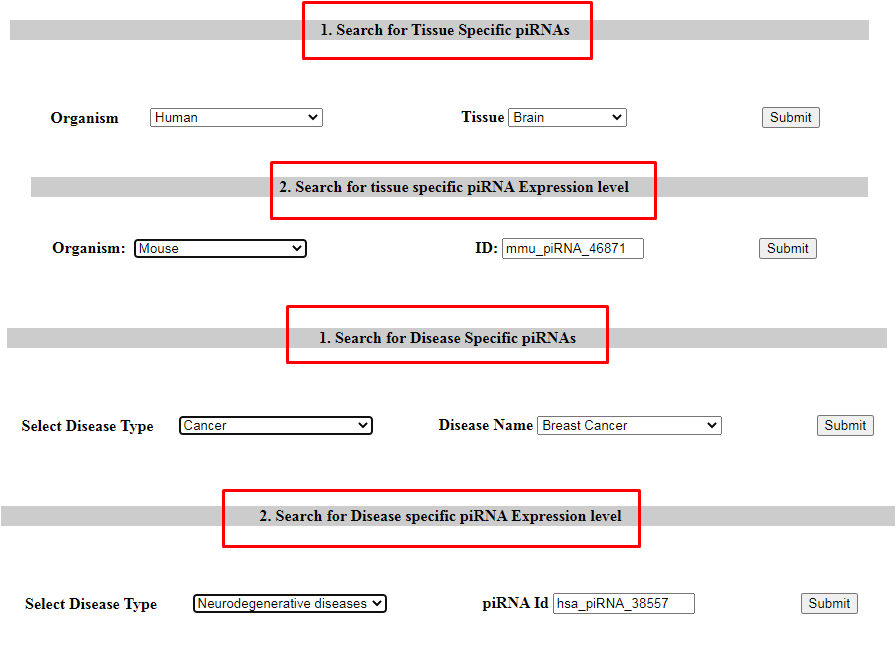
3. Search in piRNA expression
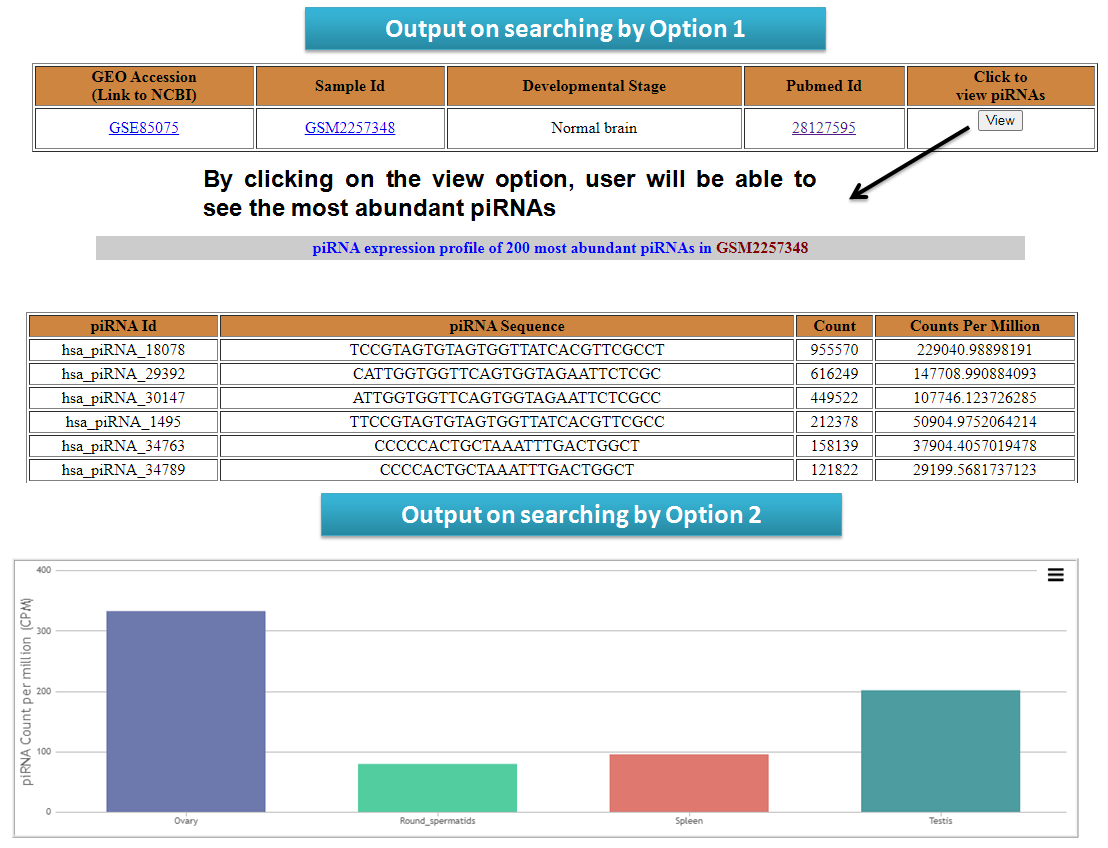
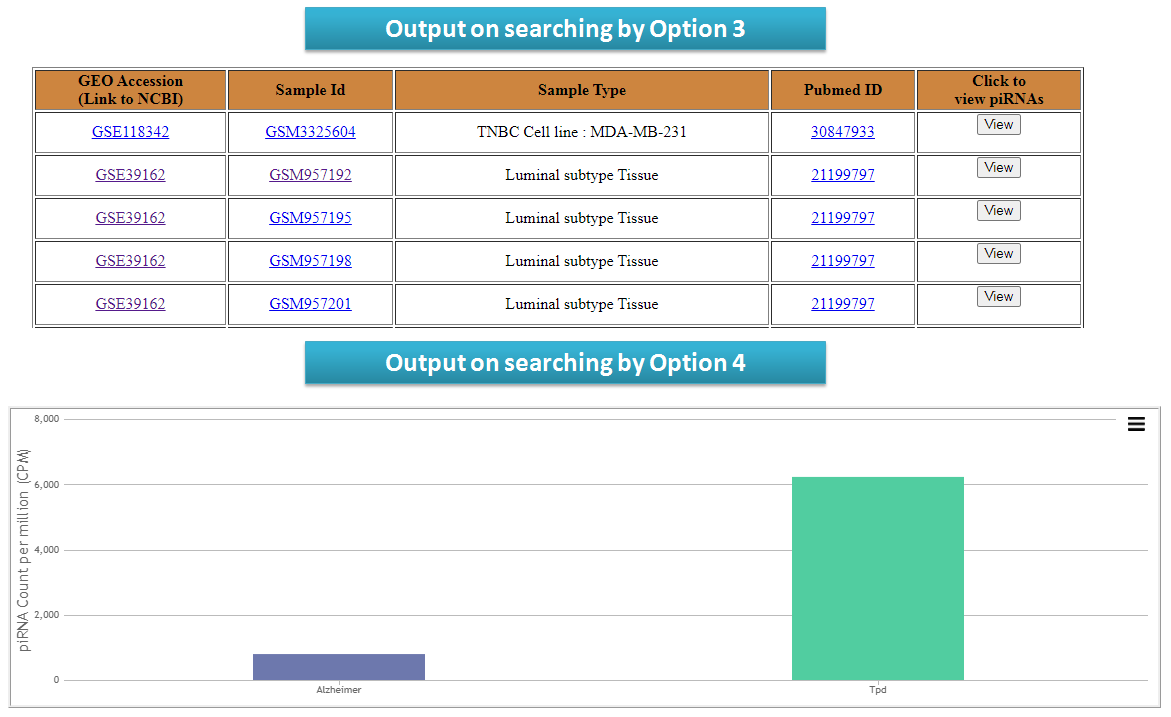
User can check piRNA expression among different tissues (Option 1 and Option 2) of corresponding species and among different disease type (Option 3 and Option 4).
View reslults for search options in piRNA Expression
|
|---|
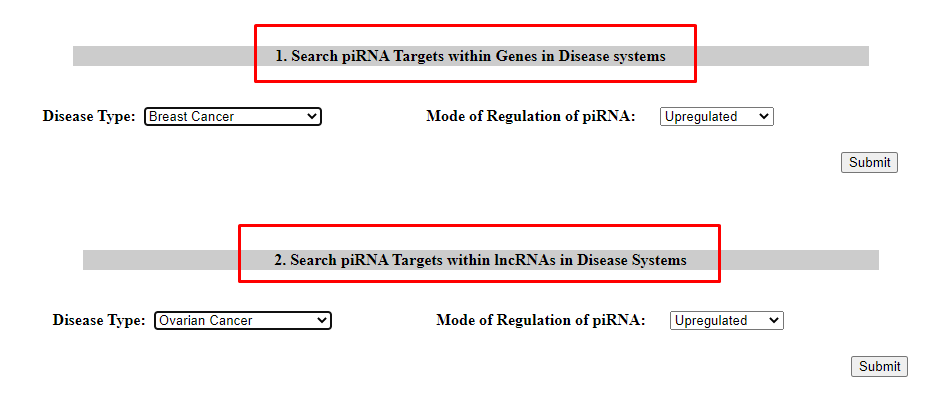
4. Search in piRNA Targets
User can check piRNA Targets in Genes (Option 1) and lncRNAs (Option 2) selecting the mode of regulation of piRNAs in a disease system.
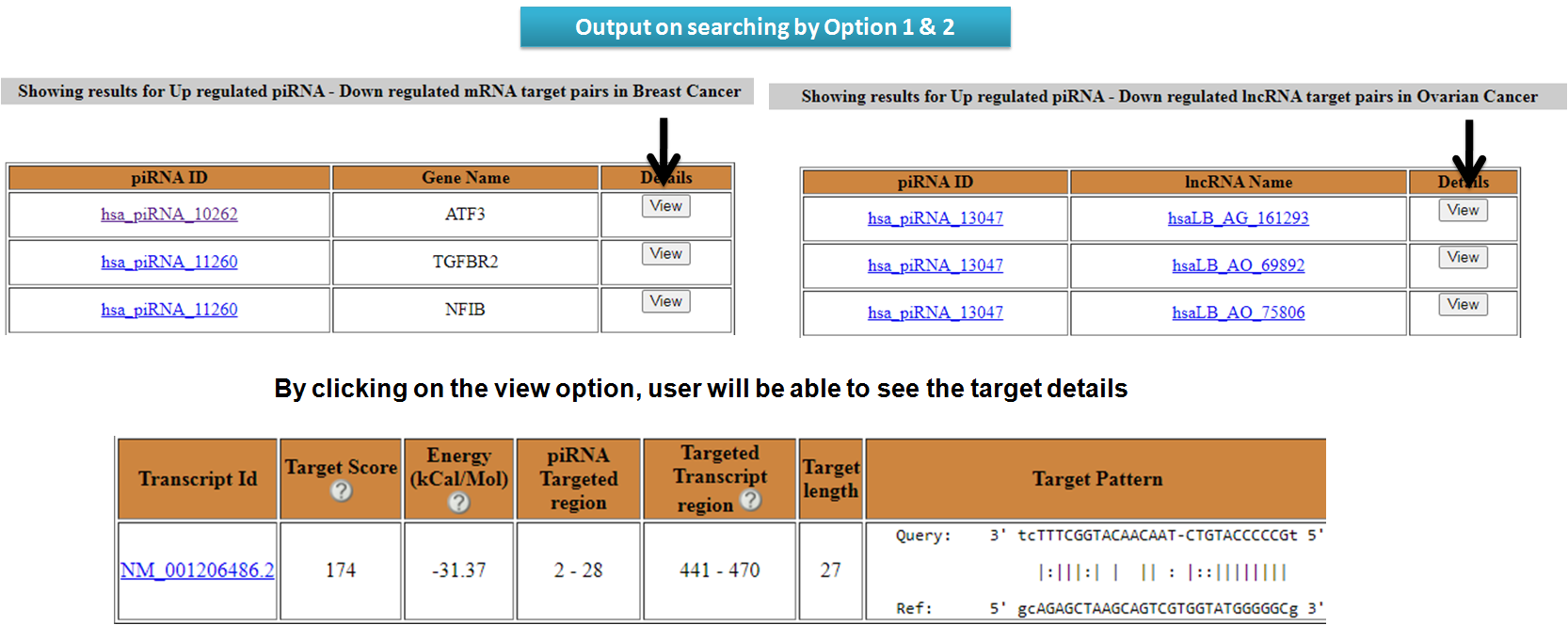
View results for search options in piRNA Targets
|
|---|
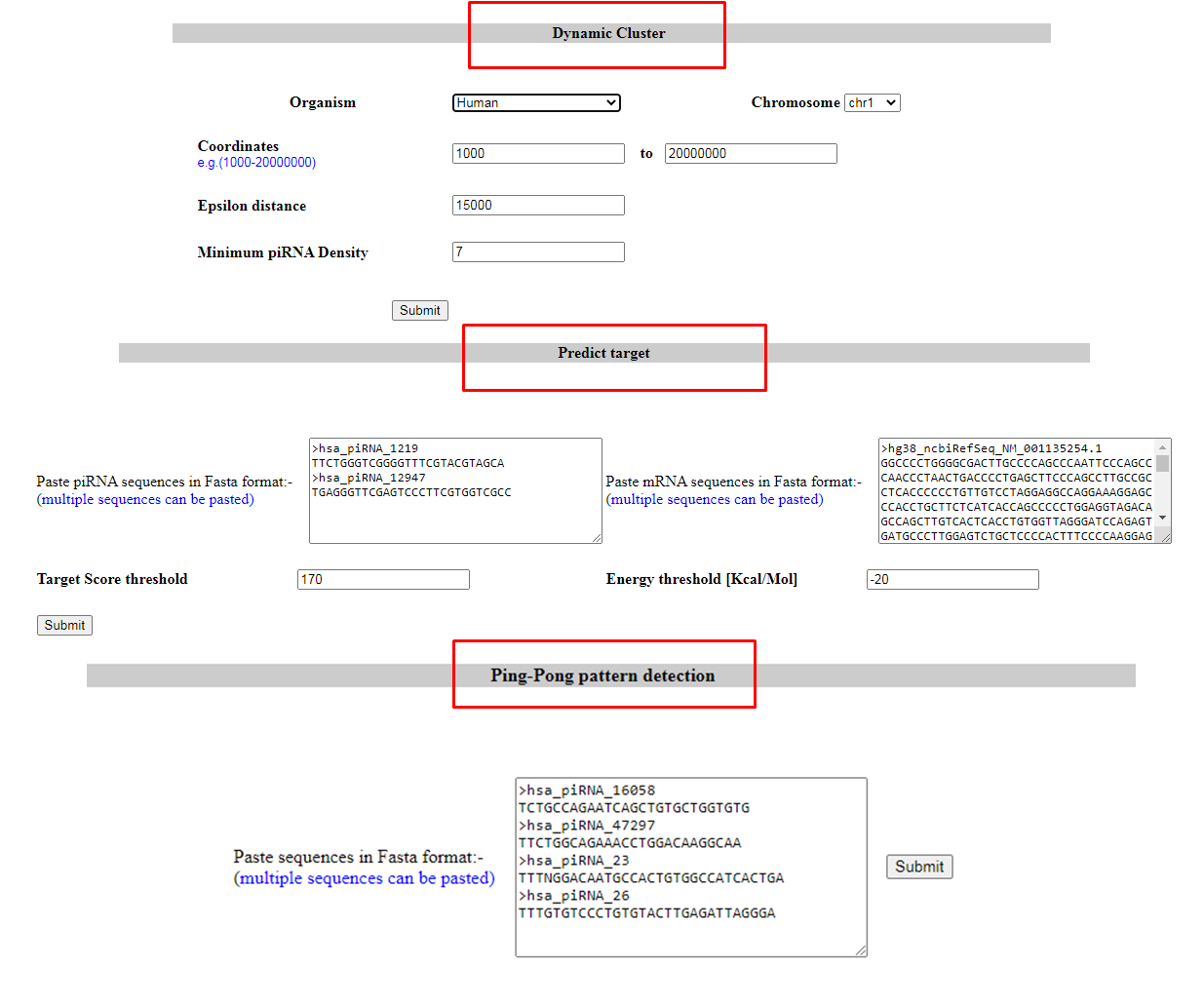
5. Tools provided
User can predict clusters and target using their own parameters with our cluster detection and target prediction (miRanda) tools. They can also check 10 nt overlapping pattern among their fasta sequences using Pingpong detection tool.